Kaggle LISH MOA Update
Hacking Away
Recap: A fellow Metis alum suggested that we enter a Kaggle competition. This project on the Method of Action of pharmeceuticals is definitely a rich sandbox to try different forms of feature processing and neural network architectures.
PCA and Clustering
Review: EDA
The input rows (one per experiment, presumably) are just labeled with a cryptic “sig_id” (presumably concealing proprietary drug chemistry…not something geochemists worry a whole lot about). The experiments are labeled to indicate whether they are experimental or control, the treatment time (24, 48, 72 hours), and a dosage flag (D1 or D2, not much help there).
The meat of the data consists of 772 columns of gene expression measurements (labeled g-0 to g-771) and 100 columns of cell viability measurements (c-0 to c-99… no clues there!). Whatever these measurements actually are, they have all been scaled from -10 to +10 (at most). If you’re curious, I’ve plotted some distributions here.
The targets are a list of 206 Methods of Action (hence the name!) that are labeled 0 or 1 for whether that treatment acted in that manner or not. For example, did the drug candidate act as a calcium channel blocker? A chelating agent? A cannabinoid receptor antagonist, perchance?
PCA
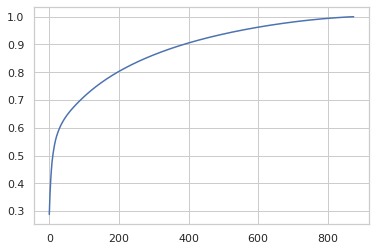
I’ve now tried a handful of strategies for preprocessing, combining feature encoding and scaling, PCA, and clustering. I’m discovering experimentally things like the dependence of PCA on the scales of the input features. In this case, if I leave treatment time at its raw values, they dominate the PCA breakdown. If, instead, I MinMaxScale them to -0.5 to 0.5 and StandardScale the gene and viability features, the PCA breakdown spreads the variation out like so:
MeanShift
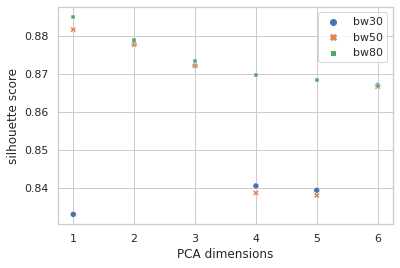
I lifted the idea of performing a clustering analysis on the data and using that as an additional set of categorical features from this notebook. Then I ran off in my own direction with it. I explored KMeans, DBSCAN, and MeanShift with varying k (1 to 10), in varying numbers of PCA dimensions (1 to 6) and it sure looked like MeanShift was at least giving the best silhouette scores, by a considerable margin in most cases. I dug down and considered the effects of setting the bandwidth at different quantiles (0.3 / 30%, 0.5 / 50%, 0.8 / 80%) for MeanShift:
How many clusters is that?
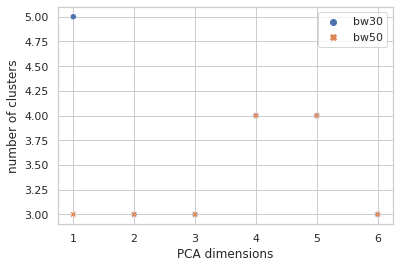
Sure seems worth the score hit to get five clusters of info with instead of four, so I went with the narrow bandwidth and… clustering in just the first PCA dimension? It looks a little quirky. Here I’m plotting the first two PCA dimensions and showing the clustering.
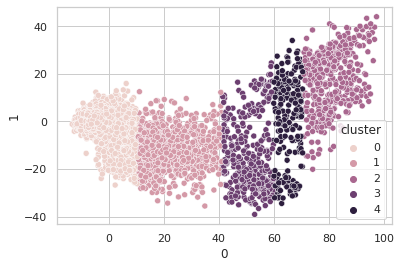
Obviously, clustering in 1-D, then plotting in 2-D, the clusters look like vertical bands. I hot encoded the cluster numbers, standard scaled the first 30 PCA dimensions, fed that to a neural network, and did indeed get a healthy bump in score once my kerastuner ground to a halt: from an uncompetitive 0.028 to a still-uncompetitive but lower 0.024. (Competitive, i.e. at least a bronze, would be below 0.0183, last I checked, and 0.01800 was just out of first place.)
Talking things over with my friend Matt, we decided to remove at least one redundant-seeming feature scaling step. I tried skipping the StandardScaling on the gene and cell features at the beginning, leaving them in the range -10 to 10 (while still MinMaxScaling the treatment times to -0.5 to 0.5 and encoding the treatment type and dose with 0 and 1), doing a PCA analysis on that, then clustering (five clusters in five PCA dimensions this time) and standard scaling these new PCA dimensions and taking 83 of them instead of 30–I have a really bad habit of trying too many things at once, but it takes Kaggle forever to grind through these notebooks and it’s hard to limit myself to one change each–when the dust had settled, I now had a score of 0.022.
Don’t get me started on trying to customize the loss function to match the competition. I’ll probably have to go back and pick that up on my own later. There are too many more interesting things to try.
–PAG
Blogging platform assembled by Jekyll, Poole, and Zach Miller of Metis.